Bioprose Blog: For several years, Los Alamos National Laboratory (LANL) scientists have been taking advantage of the molecular tools that naturally...
February 21, 2019
Author: Babetta (Babs) Marrone, Laboratory Relationship Manager for BETO programs at Los Alamos National Laboratory
Read Babetta's bio ►
Meet the other bloggers ►
Return to Bioprose blog ►
BIOPROSE BLOG
For several years, Los Alamos National Laboratory (LANL) scientists have been taking advantage of the molecular tools that naturally reside within microbial cells. Although microbes use these tools to carry out their metabolism and other life-sustaining processes, scientists can use these tools to produce fuel precursors and bioproduct building blocks. Today, researchers at LANL are further improving this process by adding a biosensor to the microbes that will use light to tell them how efficiently the product is made—thus enabling the researchers to identify cells with increased overall yield.
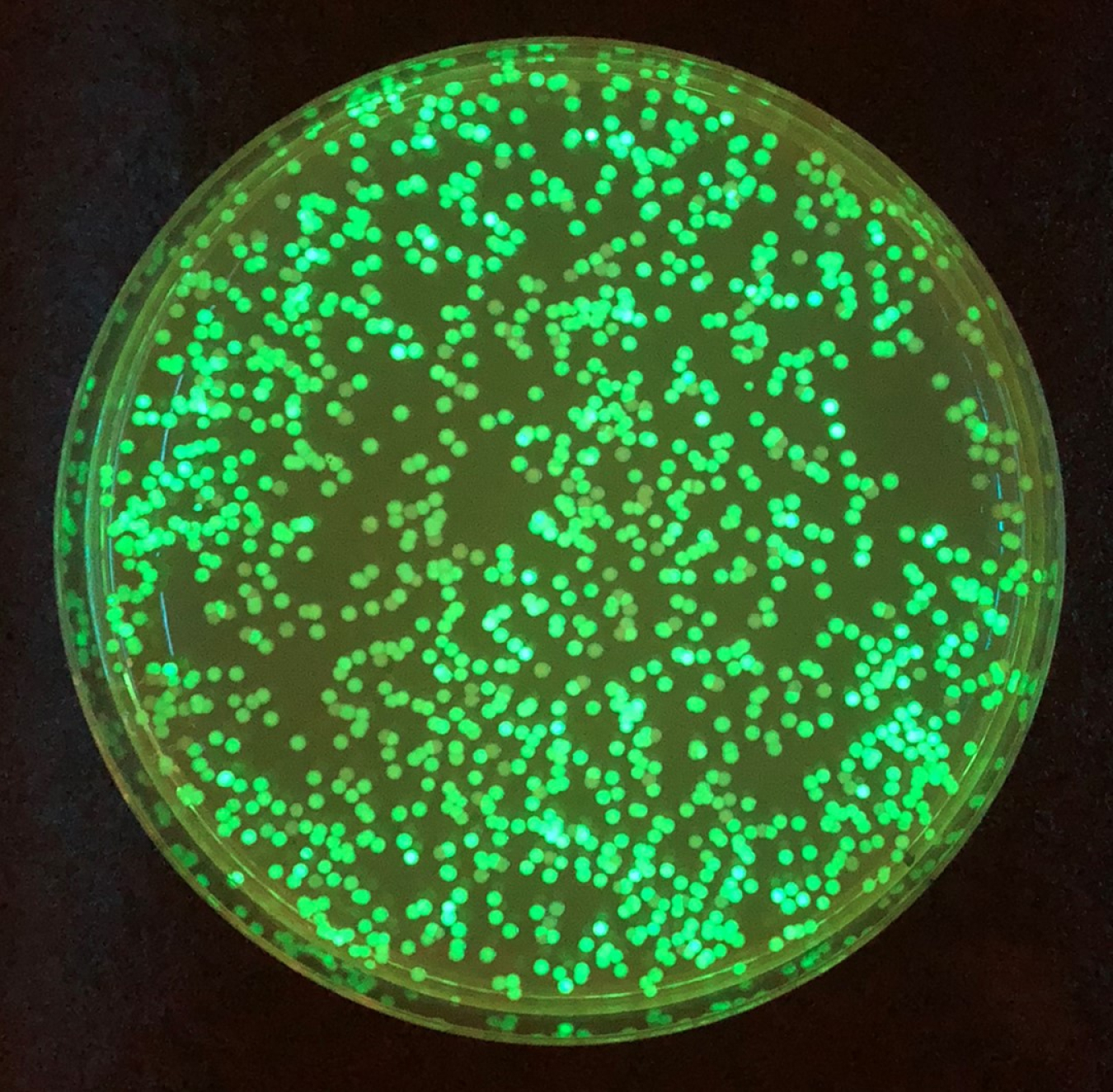
Smart microbial cell colonies "light up" when enzyme activity is high. Photo: Los Alamos National Laboratory
Babetta (Babs) L. Marrone

Dr. Babetta (Babs) L. Marrone is the former Laboratory Relationship Manager for BETO programs at Los Alamos National Laboratory (LANL). She is LANL’s Biofuels Program Manager in the Applied Energy Program Office at LANL and a Senior Scientist in the Bioscience Division.
Babs has a background in cell biology and 30+years of experience in finding innovative solutions in the biosciences. She is currently finishing a 2-year term as Senior Technologist in Residence, partnered with Procter & Gamble, in AMO’s Technologist in Residence Program, with a focus on renewable manufacturing. Marrone participates in several R&D projects sponsored by BETO. She previously led the Ultrasonic Harvesting and Extraction project for the National Alliance for Advanced Biofuels and Bioproducts (NAABB), a large algae biofuels and bioproducts consortium of over 30 institutions sponsored by BETO, from 2010-2013. She was the Director and PI of the NIH-sponsored National Flow Cytometry Resource from 2009-2014; and has led projects sponsored by NIH, LDRD, DOE-BER, DHS, and the FBI.
Meet our other bloggers ►
Return to Bioprose blog ►
More by this author
-
 Former Laboratory Relationship Manager for BETO programs at Los Alamos National Laboratory
Former Laboratory Relationship Manager for BETO programs at Los Alamos National Laboratory

